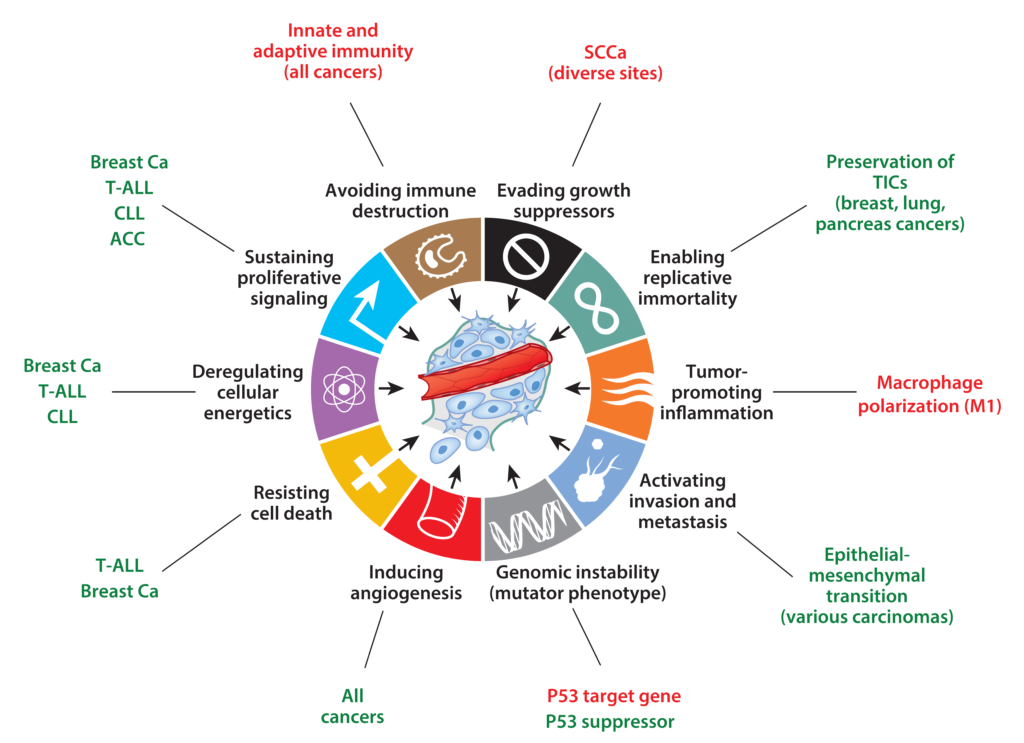
Our lab is focused on understanding the molecular pathogenesis of Notch-driven cancers such as leukemia, lymphoma, and carcinoma. We also focus on the mechanisms by which Notch1 regulates developmental processes such as cellular quiescence and differentiation, processes that contribute to tumor suppressive activities of Notch in some lineages, such as squamous cells.
Notch receptors belong to a family of structurally unique single-pass transmembrane proteins that regulate cellular differentiation during development and tissue formation in all multicellular animals. Perturbation of normal Notch signaling underlies many human diseases, particularly cancer, in which it may either be oncogenic or tumor suppressive. Normal Notch activation is triggered by binding to ligands of the DSL family expressed on neighboring cells. This induces a conformational change in a Notch juxtamembrane negative regulatory region (NRR), leading to successive cleavage by the metalloprotease ADAM10 and the intramembranous protease gamma-secretase. These cleavages release of the intracellular portion of Notch (NICD), allowing it to translocate to the nucleus and form a Notch transcription complex with the DNA-binding factor RBP-Jk/CSL and co-activators of the Mastermind family, activating target gene expression. The half-life of this complex is short, due in part to the presence of a PEST degron domain in the C-terminus of NICD.
Gain of function Notch mutations in cancer fall into two classes. One class consists of point substitution, indels, deletions, and gene rearrangements that disrupt the structure of the NRR; these mutations produce ligand-independent Notch activation and are associated with high levels of nuclear NICD. The second class consists of frameshift and nonsense mutations that result in deletion of the C-terminal PEST degron domain. These mutations stabilize NICD once it is formed. PEST mutations are sometimes found in the same allele as NRR mutations, a configuration that produces exceptionally high levels of NICD. In other instances, PEST mutations occur alone; in these tumors, receptor activation may depend on ligands expressed in the tumor microenvironment.
Other tumors, such as squamous cell carcinomas, are frequently associated with clear-cut loss of function mutations in Notch genes, indicating that Notch can function as an oncogene or a tumor suppressor in different cell states.
- What are the target genes that allow Notch to have different outcomes in different tumor types?
- What are the factors that cooperate with Notch to “condition” the epigenome to permit different transcription outputs?
- What are the ligands in the tumor microenvironment that activate Notch in tumors such as chronic lymphocytic leukemia, in which PEST mutations are common and NRR mutations rare?
- What are the biomarkers that predict responsiveness to Notch pathway inhibitors?
- Mechanistically, how does Notch activate transcription?
We are pursuing these questions in a host of model systems, using appropriate cancer cell lines, PDX models, and primary tumor cells and tissues from patients. Our work is conducted in close collaboration with the laboratory of Steve Blacklow at Harvard Medical School (structural biology, biochemistry) and Warren Pear at the University of Pennsylvania (mouse modeling).